Introduce
fastai is a modern deep learning library that provides high-level APIs to help AI programmers install deep learning models for problems like classification, segmentation … and quickly achieve good results with just as little as possible. few lines of code. In addition, thanks to its development on the Pytorch library platform, fastai also provides low-order components for researchers to develop new models, as well as fully compatible with pytorch components.
In this article, I will introduce some features of fastai and apply them to build a classification model. Let’s get started !!!
Install fastai
You can install fastai on your device with the following command:
1 2 | pip install fastai --upgrade -q |
Once installed, run the following code to import fastai and necessary libraries:
1 2 3 4 5 6 7 8 9 | <span class="token keyword">import</span> os <span class="token keyword">import</span> requests <span class="token keyword">import</span> urllib <span class="token punctuation">.</span> request <span class="token keyword">import</span> zipfile <span class="token keyword">import</span> matplotlib <span class="token punctuation">.</span> pyplot <span class="token keyword">as</span> plt <span class="token keyword">from</span> torchsummary <span class="token keyword">import</span> summary <span class="token keyword">from</span> fastai <span class="token punctuation">.</span> vision <span class="token punctuation">.</span> <span class="token builtin">all</span> <span class="token keyword">import</span> <span class="token operator">*</span> <span class="token keyword">from</span> bs4 <span class="token keyword">import</span> BeautifulSoup |
When importing fastai, some popular libraries such as numpy, pandas, matplotlib are also imported, so there is no need to re-import.
Data
I will use the data snapshot from website bacteria this . You can download data straight from the website to your computer and then unzip or if anyone uses google colab, you can use this code:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 | os <span class="token punctuation">.</span> makedirs <span class="token punctuation">(</span> <span class="token string">'dibas_zip'</span> <span class="token punctuation">)</span> os <span class="token punctuation">.</span> makedirs <span class="token punctuation">(</span> <span class="token string">'dibas_images'</span> <span class="token punctuation">)</span> url <span class="token operator">=</span> <span class="token string">'http://misztal.edu.pl/software/databases/dibas/'</span> response <span class="token operator">=</span> requests <span class="token punctuation">.</span> get <span class="token punctuation">(</span> url <span class="token punctuation">)</span> soup <span class="token operator">=</span> BeautifulSoup <span class="token punctuation">(</span> response <span class="token punctuation">.</span> text <span class="token punctuation">,</span> <span class="token string">"html.parser"</span> <span class="token punctuation">)</span> links <span class="token operator">=</span> <span class="token punctuation">[</span> tag <span class="token punctuation">[</span> <span class="token string">'href'</span> <span class="token punctuation">]</span> <span class="token keyword">for</span> tag <span class="token keyword">in</span> soup <span class="token punctuation">.</span> findAll <span class="token punctuation">(</span> <span class="token string">'a'</span> <span class="token punctuation">)</span> <span class="token punctuation">]</span> <span class="token keyword">for</span> link <span class="token keyword">in</span> links <span class="token punctuation">:</span> <span class="token keyword">if</span> <span class="token string">".zip"</span> <span class="token keyword">in</span> link <span class="token punctuation">:</span> file_name <span class="token operator">=</span> link <span class="token punctuation">.</span> partition <span class="token punctuation">(</span> <span class="token string">"/dibas/"</span> <span class="token punctuation">)</span> <span class="token punctuation">[</span> <span class="token number">2</span> <span class="token punctuation">]</span> urllib <span class="token punctuation">.</span> request <span class="token punctuation">.</span> urlretrieve <span class="token punctuation">(</span> link <span class="token punctuation">,</span> <span class="token string">'dibas_zip/'</span> <span class="token operator">+</span> file_name <span class="token punctuation">)</span> zip_ref <span class="token operator">=</span> zipfile <span class="token punctuation">.</span> ZipFile <span class="token punctuation">(</span> <span class="token string">'dibas_zip/'</span> <span class="token operator">+</span> file_name <span class="token punctuation">,</span> <span class="token string">'r'</span> <span class="token punctuation">)</span> zip_ref <span class="token punctuation">.</span> extractall <span class="token punctuation">(</span> <span class="token string">'dibas_images/'</span> <span class="token punctuation">)</span> zip_ref <span class="token punctuation">.</span> close <span class="token punctuation">(</span> <span class="token punctuation">)</span> <span class="token keyword">print</span> <span class="token punctuation">(</span> <span class="token string">"Downloaded and extracted: "</span> <span class="token operator">+</span> file_name <span class="token punctuation">)</span> |
Our data consists of 692 images:
1 2 3 4 5 6 | fns <span class="token operator">=</span> <span class="token punctuation">[</span> <span class="token punctuation">]</span> <span class="token keyword">for</span> root <span class="token punctuation">,</span> dirs <span class="token punctuation">,</span> files <span class="token keyword">in</span> os <span class="token punctuation">.</span> walk <span class="token punctuation">(</span> path <span class="token punctuation">,</span> topdown <span class="token operator">=</span> true <span class="token punctuation">)</span> <span class="token punctuation">:</span> <span class="token keyword">for</span> f <span class="token keyword">in</span> files <span class="token punctuation">:</span> fns <span class="token punctuation">.</span> append <span class="token punctuation">(</span> root <span class="token operator">/</span> Path <span class="token punctuation">(</span> f <span class="token punctuation">)</span> <span class="token punctuation">)</span> <span class="token builtin">len</span> <span class="token punctuation">(</span> fns <span class="token punctuation">)</span> <span class="token punctuation">,</span> fns <span class="token punctuation">[</span> <span class="token number">0</span> <span class="token punctuation">]</span> |
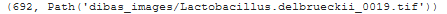
Create Dataloader
fastai provides an API for creating pytorch’s Dataloader simply and quickly
1 2 3 4 5 | dblock <span class="token operator">=</span> DataBlock <span class="token punctuation">(</span> blocks <span class="token operator">=</span> <span class="token punctuation">(</span> ImageBlock <span class="token punctuation">,</span> CategoryBlock <span class="token punctuation">)</span> <span class="token punctuation">,</span> get_y <span class="token operator">=</span> RegexLabeller <span class="token punctuation">(</span> <span class="token string">r'/(.+)_d+.tif$'</span> <span class="token punctuation">)</span> <span class="token punctuation">,</span> splitter <span class="token operator">=</span> RandomSplitter <span class="token punctuation">(</span> valid_pct <span class="token operator">=</span> <span class="token number">0.1</span> <span class="token punctuation">)</span> <span class="token punctuation">,</span> item_tfms <span class="token operator">=</span> <span class="token punctuation">[</span> Resize <span class="token punctuation">(</span> <span class="token number">512</span> <span class="token punctuation">)</span> <span class="token punctuation">]</span> <span class="token punctuation">)</span> |
The above command will return the DataBlock object. Let’s find out what each parameter is used for
block: Defines what the Dataloader will return. Since our problem is a classification problem, the Dataloader will return two things: its corresponding image and label.get_y: how to get the label from the filename. An image’s label is part of its filename. fastai provides a RegexLabeller class that uses a regular expression to separate labels from filenames. For example:
1 2 | RegexLabeller(r'/(.+)_d+.tif$')('dibas_images/Lactobacillus.delbrueckii_0019.tif') |
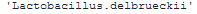
splitter: dividing dataset into 2 sets of train / validationitem_tfms: Because the image is of different sizes, we need to resize it to the same size before we can pack in batches.
After having Datablock, just run:
1 2 | dls <span class="token operator">=</span> dblock <span class="token punctuation">.</span> dataloaders <span class="token punctuation">(</span> source <span class="token operator">=</span> fns <span class="token punctuation">,</span> bs <span class="token operator">=</span> <span class="token number">16</span> <span class="token punctuation">)</span> |
along with the following parameters: data source (list of image files) and batch size. The above method will return a Dataloaders object. As the name implies, Dataloaders includes many Dataloaders (1 train and 1 validation). People can index into dls to access the dataloaders: dls [0], dls [1].
We can check how many classes the data set has:
1 2 | print(dls.vocab) |
Training
Model training is handled by the Learner class. With classification problem you can create Learner by function cnn_learner :
1 2 | learn <span class="token operator">=</span> cnn_learner <span class="token punctuation">(</span> dls <span class="token punctuation">,</span> resnet50 <span class="token punctuation">,</span> metrics <span class="token operator">=</span> accuracy <span class="token punctuation">)</span> |
The parameters include:
- Dataloaders
- CNN architecture. Here I use Resnet50 but people can use CNN pretrain networks available on torchvision
- List of metrics
When using the pretrain model, learner will automatically add some Linear classes at the end of the CNN section
1 2 | learn <span class="token punctuation">.</span> model |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | (1): Sequential( (0): AdaptiveConcatPool2d( (ap): AdaptiveAvgPool2d(output_size=1) (mp): AdaptiveMaxPool2d(output_size=1) ) (1): Flatten(full=False) (2): BatchNorm1d(4096, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True) (3): Dropout(p=0.25, inplace=False) (4): Linear(in_features=4096, out_features=512, bias=False) (5): ReLU(inplace=True) (6): BatchNorm1d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True) (7): Dropout(p=0.5, inplace=False) (8): Linear(in_features=512, out_features=33, bias=False) ) |
By default, the weight of the CNN will be frozen and not updated during the training.
The training model is very simple:
1 2 | learn <span class="token punctuation">.</span> fit_one_cycle <span class="token punctuation">(</span> <span class="token number">8</span> <span class="token punctuation">,</span> <span class="token number">1e</span> <span class="token operator">-</span> <span class="token number">3</span> <span class="token punctuation">)</span> |
fit_one_cycle(8, 1e-3) will train model for 8 epoch using a 1-cycle policy . If you don’t want to use the learning rate scheduler, you can use the fit method
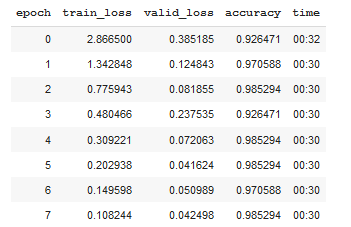
After just 8 epochs, the accuracy will reach 98.5%. Now we will break the ice to train CNN:
1 2 3 | learn.unfreeze() learn.fit_one_cycle(5, 1e-7) |
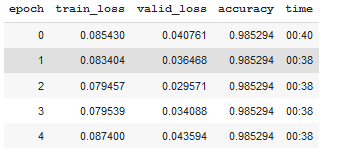
To see the results of the model, everyone can run learn.show_results()
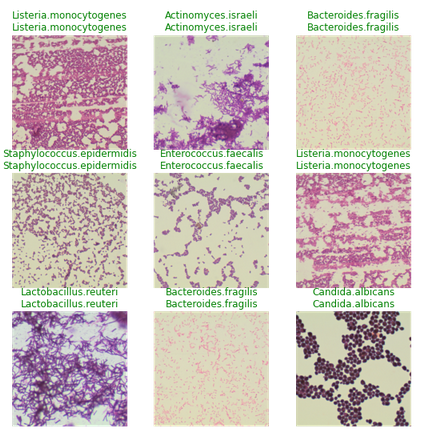
The whole process from loading the data to the train model takes less than 10 lines of code
1 2 3 4 5 6 7 8 9 10 | dblock <span class="token operator">=</span> DataBlock <span class="token punctuation">(</span> blocks <span class="token operator">=</span> <span class="token punctuation">(</span> ImageBlock <span class="token punctuation">,</span> CategoryBlock <span class="token punctuation">)</span> <span class="token punctuation">,</span> get_y <span class="token operator">=</span> RegexLabeller <span class="token punctuation">(</span> <span class="token string">r'/(.+)_d+.tif$'</span> <span class="token punctuation">)</span> <span class="token punctuation">,</span> splitter <span class="token operator">=</span> RandomSplitter <span class="token punctuation">(</span> valid_pct <span class="token operator">=</span> <span class="token number">0.1</span> <span class="token punctuation">)</span> <span class="token punctuation">,</span> item_tfms <span class="token operator">=</span> <span class="token punctuation">[</span> Resize <span class="token punctuation">(</span> <span class="token number">512</span> <span class="token punctuation">)</span> <span class="token punctuation">]</span> <span class="token punctuation">)</span> dls <span class="token operator">=</span> dblock <span class="token punctuation">.</span> dataloaders <span class="token punctuation">(</span> source <span class="token operator">=</span> fns <span class="token punctuation">,</span> bs <span class="token operator">=</span> <span class="token number">16</span> <span class="token punctuation">)</span> learn <span class="token operator">=</span> cnn_learner <span class="token punctuation">(</span> dls <span class="token punctuation">,</span> resnet50 <span class="token punctuation">,</span> metrics <span class="token operator">=</span> accuracy <span class="token punctuation">)</span> learn <span class="token punctuation">.</span> fit_one_cycle <span class="token punctuation">(</span> <span class="token number">8</span> <span class="token punctuation">,</span> <span class="token number">1e</span> <span class="token operator">-</span> <span class="token number">3</span> <span class="token punctuation">)</span> learn <span class="token punctuation">.</span> unfreeze <span class="token punctuation">(</span> <span class="token punctuation">)</span> learn <span class="token punctuation">.</span> fit_one_cycle <span class="token punctuation">(</span> <span class="token number">5</span> <span class="token punctuation">,</span> <span class="token number">1e</span> <span class="token operator">-</span> <span class="token number">7</span> <span class="token punctuation">)</span> |
Epilogue
Above, I have instructed you to install bacteria classification model with 98.5% accuracy using the fastai library. With less than 10 lines of code, we have passed the 97% SOTA results on this dataset (you can check here ). If you find this article useful, please leave me an upvode. Thank you all for your interest and see you in the next articles.
